For the first time, scientists have shown that gut bacteria differences are associated with later development of juvenile idiopathic arthritis, a debilitating rheumatic childhood disease, and that these differences are present years before the disease is diagnosed.
The research team, which includes scientists from the University of Florida and researchers in Sweden, made this discovery by analyzing stool samples from one-year-old children in a long-term study called All Babies in Southeast Sweden. The scientists compared bacteria found in children who went on to develop juvenile idiopathic arthritis with those who did not.
“Our work suggests that an imbalance in microbes, especially the increased prevalence of several proinflammatory bacterial species, could serve as a potential indicator of future disease risk,” said Angelica Ahrens, co-first author of the study and a postdoctoral associate in the UF/IFAS department of microbiology and cell science.
Juvenile idiopathic arthritis, or JIA, which is also called childhood arthritis and pediatric rheumatic disease, is an autoimmune disease marked by inflammation of the joints and sometimes other parts of the body. Children with JIA experience pain, swelling, stiffness and other symptoms that make daily activities challenging. Current treatments for JIA include anti-inflammatory drugs and steroid injections.
“These treatments can help control inflammation and reduce symptoms, but they are not without their drawbacks and they do not cure the disease. There is a need to find alternative approaches, and the bacteria found in gastrointestinal tract may be a promising place to start,” said Dr. Erik Kindgren, co-first author of the study and a pediatrician at Skaraborg Hospital in Sweden who treats children with JIA.
While other studies have shown microbial differences in children already diagnosed with JIA, this study is the first to demonstrate that these differences are present years before children first show symptoms of the disease.
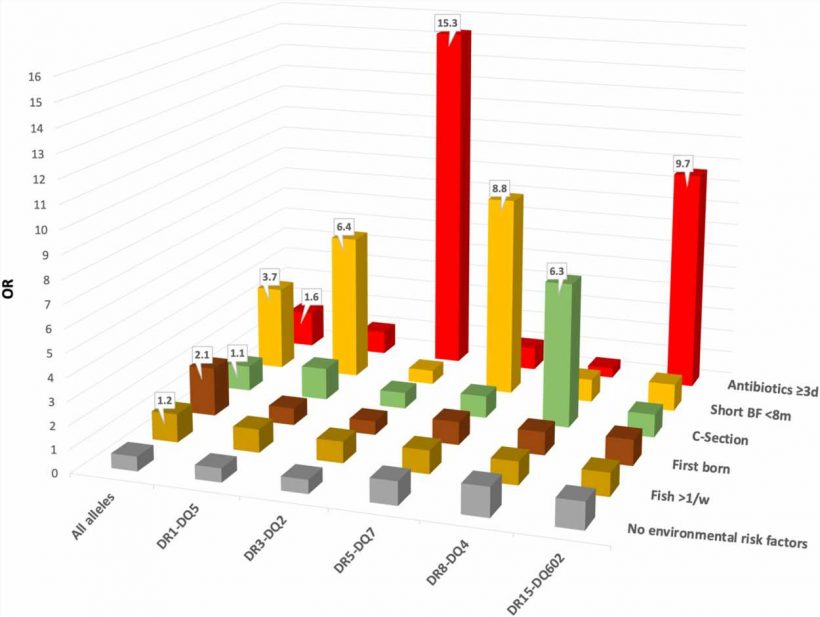
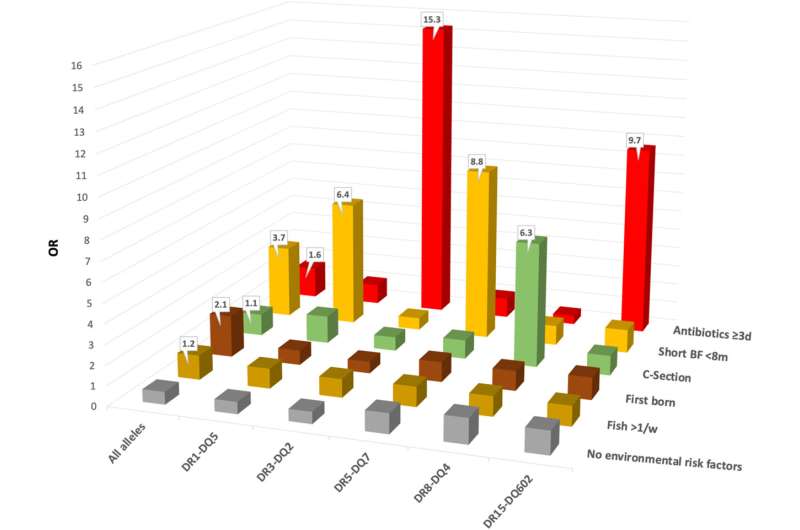
The study found that children with gut bacteria known to cause inflammation were nearly seven times more likely to develop JIA. The researchers also found that bacteria known to promote a healthy gut lining were absent or reduced in children who later developed the disease. These trends held true even when the researchers controlled for factors already associated with the disease, such as being breastfed for shorter periods or early exposure to antibiotics.
The scientists say the findings are a first step toward understanding what causes JIA—the term “idiopathic” in the name means the cause of the condition is unknown.
“Looking ahead, this line of discovery could lead to the development of screening tools in early pediatric wellness visits. By constructing risk profiles and implementing targeted interventions and preventative measures to reduce those risks, we may be able to prevent disease onset in some people,” Ahrens said.
First, though, researchers will need to understand how the bacteria identified in the study contribute to the disease.
“Functionally, what are these bacteria doing in the body that leads to this disease? That’s what we need to investigate next,” Ahrens said.
The current study only looked at a snapshot of the gut microbiome at one year of age, so future work may investigate how the gut microbiome of children with JIA progresses over time.
The study’s authors—which also include Eric Triplett, chair of the UF/IFAS department of microbiology and cell science, and Dr. Johnny Ludvigsson, senior professor at Linköping University and both founder and leader of the All Babies in Southeast Sweden study—plan to continue their trans-Atlantic collaboration by examining microbial differences associated with other conditions that appear in childhood.
More information:
Erik Kindgren et al, Infant gut microbiota and environment associate with juvenile idiopathic arthritis many years prior to disease onset, especially in genetically vulnerable children, eBioMedicine (2023). DOI: 10.1016/j.ebiom.2023.104654
Journal information:
EBioMedicine
Source: Read Full Article