India’s surge in SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) infections are linked to the new variants B.1.617 and B.1.618, with mutated spike proteins. In recent months, it has caused a devastating second wave of the coronavirus disease 2019 (COVID-19) pandemic. According to the World Health Organization (WHO), it is reported that from 3 January 2020 to 17 May 2021, there have been over 25 million confirmed cases of COVID-19 with over 274 thousand deaths.
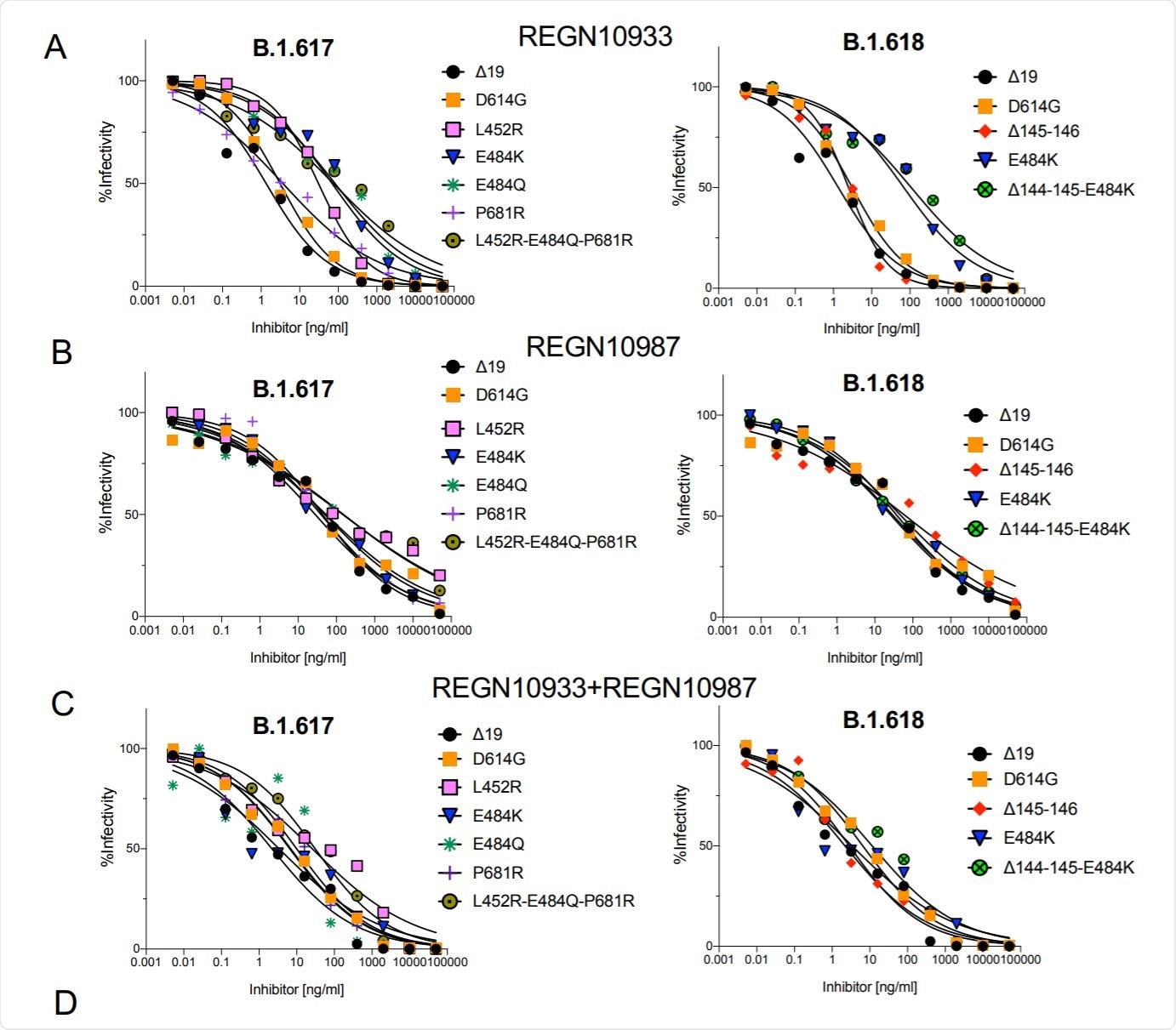
Neutralization of B.1.617 and B.1.618 spike protein variants by REGN10933 and REGN10987. Image Credit: https://www.biorxiv.org/content/10.1101/2021.05.14.444076v1.full.pdf
The mutations in these variants may contribute to the increased transmissibility of the virus, and could potentially result in re-infection or resistance to the vaccine-elicited antibodies. The mutations are driven by selective pressure for increased affinity for its receptor, ACE2 (angiotensin-converting enzyme), and escape from neutralizing antibodies. This raises concern over the fitness of the Indian SARS-CoV-2 variants and their ability to escape the vaccine-elicited immune response.
In this context, researchers from the NYU Grossman School of Medicine, New York, USA, tested the neutralization of B.1.617 and B.1.618 SARS-CoV-2 variant spike proteins and determined their resistance to neutralization by convalescent sera, vaccine-elicited antibodies, and therapeutic monoclonal antibodies.
To achieve this they generated lentiviruses pseudotyped by the variant proteins. They found that these viruses with B.1.617 and B.1.618 spike proteins were neutralized with a 2-5-fold decrease in titer by convalescent sera and vaccine-elicited antibodies.
They observed a modest neutralization resistance to the vaccine-elicited antibodies. This is good news as it suggests that the current vaccines will protect against the B.1.617 and B.1.618 42 variants. This study, led by Professor Nathaniel R. Landau, is recently posted on the bioRxiv* server.
Our results lend confidence that current vaccines will provide protection against variants identified to date,”
The researchers also found that the resistance was caused by the L452R, E484Q, and E484K mutations. Further, they reported that the variants were partially resistant to REGN10933, which is one of the two mAbs constituting the Regeneron COV2 therapy (casirivimab (REGN10933) with imdevimab (REGN10987), for the treatment of mild-to-moderate COVID-19).
The B.1.617 encodes a spike protein with the mutations L452R, E484Q, D614G, and P681R while the B.1.618 spike has mutations Δ145-146, E484K, and D614G. The B.1.617 variant spike protein contains L452R and E484Q mutations in the RBD in addition to D614G and the P681R mutation near the proteolytic processing site and the B.1.618 spike has E484K in the RBD in addition to D614G and the N-terminal deletion Δ145-146.
The researchers generated the lentiviral virions, expressing the spike proteins at a level similar to that of wild-type D614G. They also tested the infectivity of the virus, reporting that the B.1.617 spike protein (L452R/E484Q/P681R) was >2-fold increase in infectivity while B.1.618 was similar to wild-type D614G.
Significantly, they found that the increased infectivity of the B.1.617 spike was attributed to L452R mutation, which caused a 3.5-fold increase in infectivity and, in combination with E484Q caused a 3-fold increase. Other point mutations had an insignificant effect on the infectivity.
Both variants B.1.617 and B.1.618 have increased affinity for ACE2 and the researchers found that both are partially resistant to the monoclonal antibodies. They discussed the mutations, the expressed proteins, and the subsequent effect on binding and infection.
In this study, the researchers reported that the virus variants B.1.617 and B.1.618 spike were partially resistant to neutralization, with an average 3.9-fold and 2.7-fold decrease in IC50 for convalescent sera and antibodies elicited by Pfizer and Moderna mRNA vaccines, respectively. The neutralization resistance was mediated by the L452R, E484Q, and E484K mutations. The resistance of these variants is similar to the previous variants.
Even with the 3-4-fold decrease in neutralization titer of vaccine-elicited antibodies, average titers were around 1:500, a titer well above that found in the sera of individuals who have recovered from infection with earlier unmutated viruses.”
Significantly, this study reassures that the vaccinated individuals will remain protected against the B.1.617 and B.1.618 variants.
Commenting on the other vaccines, the researchers said, “The analyses in this study were restricted to the mRNA-based vaccines but there is no reason to believe that vector-based vaccines such as that of Johnson and Johnson that express a stabilized, native, full-length spike protein would be different with regarding antibody neutralization of virus variants.”
- The Spike Proteins of SARS-CoV-2 B.1.617 and B.1.618 Variants Identified in India Provide Partial Resistance to Vaccine-elicited and Therapeutic Monoclonal Antibodies. Takuya Tada, Hao Zhou, Belinda M Dcosta, Marie I Samanovic, Mark J Mulligan, Nathaniel R Landau bioRxiv 2021.05.14.444076; doi: https://doi.org/10.1101/2021.05.14.444076
Posted in: Medical Science News | Medical Research News | Disease/Infection News | Healthcare News
Tags: ACE2, Angiotensin, Antibodies, Antibody, Coronavirus, Coronavirus Disease COVID-19, Enzyme, Immune Response, Medicine, Mutation, Pandemic, Protein, Receptor, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Vaccine, Virus

Written by
Dr. Ramya Dwivedi
Ramya has a Ph.D. in Biotechnology from the National Chemical Laboratories (CSIR-NCL), in Pune. Her work consisted of functionalizing nanoparticles with different molecules of biological interest, studying the reaction system and establishing useful applications.
Source: Read Full Article