A recent study posted to the bioRxiv* preprint server demonstrated that the latest severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron sublineages evade immunity imparted by the prior subvariants.
 Study: BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Image Credit: NIAID
Study: BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Image Credit: NIAID
Background
The SARS-CoV-2 Omicron (B.1.1.529) variant's recent emergence and worldwide distribution have presented a potent threat to the effectiveness of CoV disease 2019 (COVID-19) vaccines and neutralizing antibodies (NAbs) therapeutics. Omicron can produce significant NAb evasion because of numerous mutations in the spike (S) protein, including its N-terminal domain (NTD) and receptor-binding domain (RBD).
Most importantly, the newly emerged SARS-CoV-2 Omicron BA.5, BA.4, BA.2.13, and BA.2.12.1 subvariants include the L452 mutation and exhibit possible significant transmissibility than the Omicron BA.2 sublineage. The capacity of the novel mutants to connect to receptors and evade the immune system necessitates rapid exploration, particularly into the impact of L452 substitutions.
About the study
In the present study, the scientists estimated the recently emerged SARS-CoV-2 Omicron sublineages with the previous Omicron subvariants. The authors purified and expressed the prefusion-stabilized trimeric ectodomains of the Omicron BA.4/BA.5, BA.2.13, BA.3, 24, BA.2, BA.1, and BA.2.12.1 Ss to thoroughly analyze the functional and structural characteristics of the Omicron sublineages.
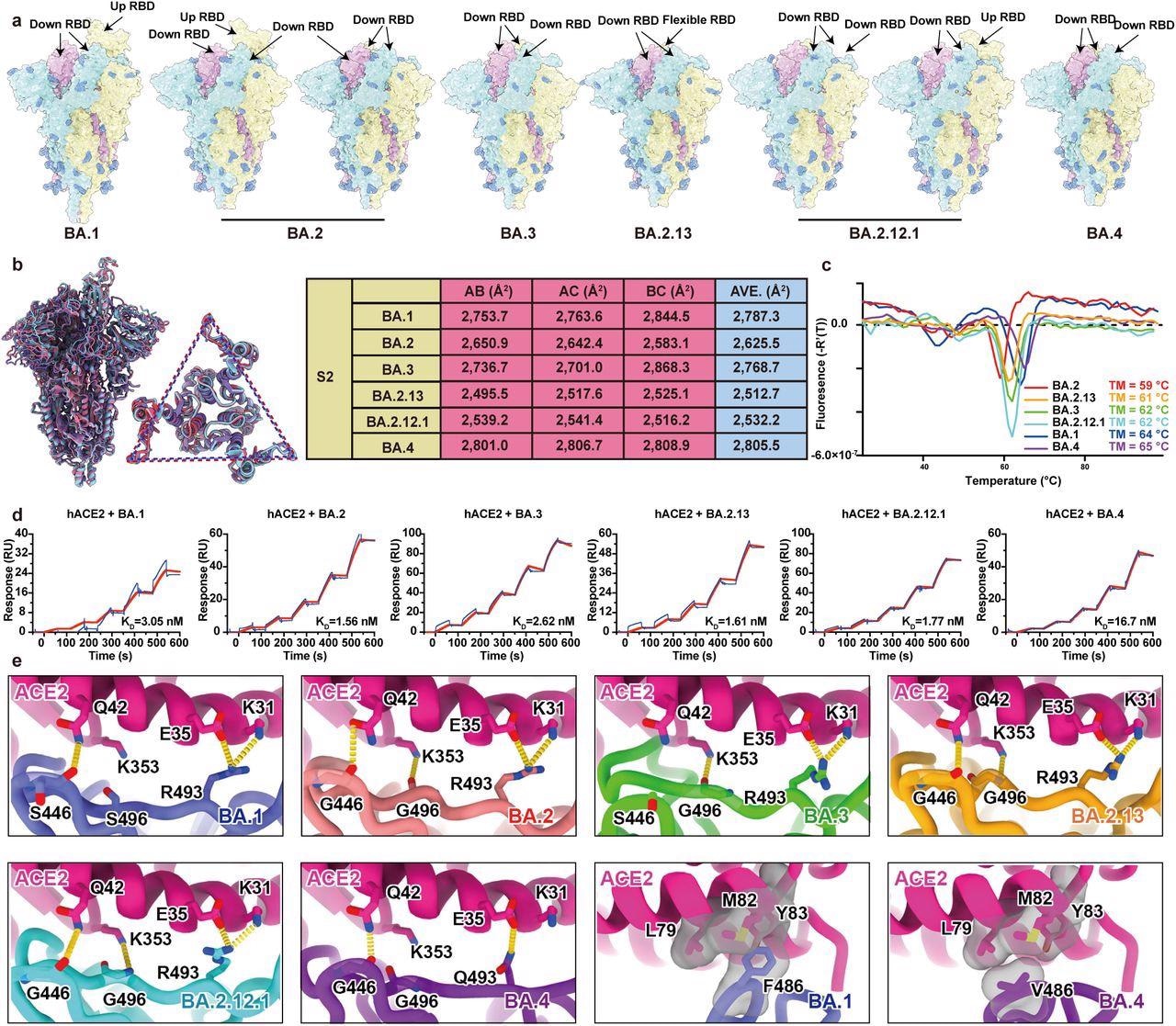
Structural characteristics of Omicron subvariants. a, Surface representation for structures of S trimer of BA.1, BA.2, BA.3, BA.2.13, BA.2.12.1 and BA.4 variants. b, Structural interpretation and functional verification for the stability of S protein for BA.1, BA.2, BA.3, BA.2.13, BA.2.12.1 and BA.4 variants. The superimposed structures for S protein and the S2 domains of BA.1 (purple), BA.2 (red) and BA.4 (blue) are shown on the left. The binding surface areas between S2 subunits of all variants are calculated in the table on the right. c, Thermoflour analysis for these Omicron variants. Curves for each Omicron variant are colored in rainbow (BA.1: blue; BA.2: red; BA.3: green; BA.2.13: orange; BA.2.12.1: cyan; BA.4: purple). d, Binding affinities of S-trimers of Omicron variants to hACE2 measured by SPR. e, Details of interactions between hACE2 and RBD of Omicron variants. Structures of the RBD from Omicron variants and hACE2 are shown as ribbons. Color scheme is same as b.
a, Surface representation for structures of S trimer of BA.1, BA.2, BA.3, BA.2.13, BA.2.12.1 and BA.4 variants. b, Structural interpretation and functional verification for the stability of S protein for BA.1, BA.2, BA.3, BA.2.13, BA.2.12.1 and BA.4 variants. The superimposed structures for S protein and the S2 domains of BA.1 (purple), BA.2 (red) and BA.4 (blue) are shown on the left. The binding surface areas between S2 subunits of all variants are calculated in the table on the right. c, Thermoflour analysis for these Omicron variants. Curves for each Omicron variant are colored in rainbow (BA.1: blue; BA.2: red; BA.3: green; BA.2.13: orange; BA.2.12.1: cyan; BA.4: purple). d, Binding affinities of S-trimers of Omicron variants to hACE2 measured by SPR. e, Details of interactions between hACE2 and RBD of Omicron variants. Structures of the RBD from Omicron variants and hACE2 are shown as ribbons. Color scheme is same as b.
Surface plasmon resonance (SPR) was used to determine the binding affinity among S-trimers of the Omicron variants and human angiotensin-converting enzyme 2 (hACE2). The researchers used the D614G mutant and Omicron BA.4/BA.5, BA.1, BA.2.13, BA.1.1, BA.2, BA.2.12.1, and BA.3 subvariants in pseudovirus neutralization experiments against plasma from vaccinated COVID-19 convalescents, BA.1 convalescents, and three-dose vaccinated subjects. The goal was to determine how well the newly discovered Omicron subvariants might evade neutralization.
The team analyzed how therapeutic antibodies against Omicron sublineages differed in their neutralizing activity. The epitope distribution, escape mutation patterns, and Omicron subvariant neutralization efficiency of 1640 RBD-targeted NAbs, like 614 derived from BA.1 convalescents, were evaluated to discover the underpinning evasion mechanism. High-throughput yeast-display-based deep mutational scanning (DMS) evaluations spanning all potential single residue changes in SARS-CoV-2 wild-type (WT) RBD background were performed to learn more about the epitope distribution of NAbs evoked by post-vaccination BA.1 infection. Further, the authors tested the NAbs' ability to neutralize BA.4/BA.5, BA.2.12.1, and BA.2.13, and the main Omicron variants: BA.3, BA.1, BA.2 17, and BA.1.1.
Results
The study results showed that the Omicron BA.2 sublineages, such as BA.2.13 and BA.2.12.1, had higher ACE2-binding affinities than BA.1, but BA.5/BA.4 has the poorest receptor-binding ability due to R493Q and F486V reversion using structural comparisons. BA.5/BA.4 and BA.2.12.1 relative to BA.2 showed higher neutralization evasion from the plasma of three-dose vaccinees and, most notably, from vaccinated BA.1 convalescents.
Surprisingly, BA.1 infection after COVID-19 vaccination evoked antibodies that neutralized both BA.1 and WT and mostly recalled WT-generated humoral memory. Furthermore, these cross-reactive NAbs were substantially abundant with non-ACE2-competing epitopes. Oddly, the bulk of these NAbs, including L452M (BA.2.13), R346K (BA.1.1), L452R (BA.4/BA.5), and L452Q (BA.2.12.1), were weakened by L452 and R346 substitutions. This inference implied that L452 and R346K mutations arose under the immunological stress of Omicron convalescents.
Nevertheless, due to the high sensitivity to E484, N440, N501, and K417 mutations, BA.1 infection could also generate new clones of BA.1-specific antibodies that could neutralize BA.1 yet not the SARS-CoV-2 WT strain. Interestingly, because of F486V and D405N, these NAbs were mostly evaded by BA.2 subvariants and BA.5/BA.4, which have narrow neutralization breadths. In terms of therapeutic NAbs, CoV2-2130 (Cilgavimab) and LY-CoV1404 (Bebtelovimab) could still neutralize BA.5/BA.4 and BA.2.12.1, albeit the R408S, S371F, and D405N mutations harbored by the BA.5/BA.2/BA.4 sublineages would render most expansive sarbecovirus NAbs useless.
These data imply that various recent Omicron sublineages could develop mutations to avoid humoral immunity induced by the Omicron BA.1 subvariant infection. Omicron's continued advancement presents significant hurdles to SARS-CoV-2 herd immunity, suggesting that BA.1-derived COVID-19 vaccine boosters might not be the best option for obtaining broad protection.
Conclusions
The study findings demonstrated that Omicron was constantly evolving under immunological strain and explained why L452 and R346K (BA.1.1) substitutions emerged. Unlike when Omicron originally arose, Omicron subvariants have begun to address humoral immunity generated by Omicron, such as humoral immunity established by Omicron infection after vaccination. This presents a substantial threat to the presently developed herd immunity acquired by BA.1/BA.2 infection and WT-based vaccination.
Likewise, this finding suggested that an antigen based on Omicron BA.1 might not be the best antigen for triggering broad-spectrum protection towards arising Omicron subvariants. According to the authors, the detailed information presented in this paper is crucial for the development of broad-spectrum antibodies and vaccines against sarbecoviruses.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- BA.2.12.1, BA.4, and BA.5 escape antibodies elicited by Omicron infection; Yunlong Richard Cao, Ayijiang Yisimayi, Fanchong Jian, Weiliang Song, Tianhe Xiao, Lei Wang, Shuo Du, Jing Wang, Qianqian Li, Xiaosu Chen, Peng Wang, Zhiying Zhang, Pulan Liu, Ran An, Xiaohua Hao, Yao Wang, Jing Wang, Rui Feng, Haiyan Sun, Lijuan Zhao, Wen Zhang, Dong Zhao, Jiang Zheng, Lingling Yu, Can Li, Na Zhang, Rui Wang, Xiao Niu, Sijie Yang, Xuetao Song, Linlin Zheng, Zhiqiang Li, Qingqing Gu, Fei Shao, Weijin Huang, Ronghua Jin, Zhongyang Shen, Youchun Wang, Xiangxi Wang, Junyu Xiao, Xiaoliang Sunney Xie. bioRxiv preprint 2022. DOI: https://doi.org/10.1101/2022.04.30.489997, https://www.biorxiv.org/content/10.1101/2022.04.30.489997v1
Posted in: Medical Research News | Disease/Infection News
Tags: ACE2, Angiotensin, Angiotensin-Converting Enzyme 2, Antibodies, Antigen, binding affinity, Coronavirus, Coronavirus Disease COVID-19, covid-19, Enzyme, Immune System, immunity, Mutation, Omicron, Protein, Pseudovirus, Receptor, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Stress, Syndrome, Therapeutics, Vaccine, Yeast

Written by
Shanet Susan Alex
Shanet Susan Alex, a medical writer, based in Kerala, India, is a Doctor of Pharmacy graduate from Kerala University of Health Sciences. Her academic background is in clinical pharmacy and research, and she is passionate about medical writing. Shanet has published papers in the International Journal of Medical Science and Current Research (IJMSCR), the International Journal of Pharmacy (IJP), and the International Journal of Medical Science and Applied Research (IJMSAR). Apart from work, she enjoys listening to music and watching movies.
Source: Read Full Article